Past Research
Magnetic Resonance and Biological Applications
The initial research in my group focused on Magnetic Resonance and the applications of electron paramagnetic resonance, EPR, to the study of biological processes. In particular, my group has applied relaxation theory to explaining the EPR lineshapes and relaxation rates of nitroxide molecules undergoing dynamics generally associated with proteins and nucleic acids. Relaxation theory explains the resonance response due to the dynamics of the environment on the orientation of an EPR active probe as an example of non-equilibrium, irreversible quantum-statistical dynamics. As one example, shown to the right, we developed visualization methods to show how membrane-binding proteins orient on membrane surfaces and penetrate into the membrane core. We also developed a structural model to describe ways in which DNA base pairs stack; and how that stacking modifies base-pair dynamics and cooperative transitions that may involve many base pairs simultaneously.
Magnetic Resonance Methodology
We have also developed a model for fully accounting for the line broadening of EPR-active, nitroxide spin-labels due to spectrometer effects and relaxation processes. This model has been used extensively by EPR based imaging techniques. We have also developed direct methods to simulate MR-active probe images, and methods to determine the velocities and directions of blood flow in brains (which has led to a patent). We have also developed fluorescent imaging methods to visualize organelles in cells (which has also led to two patents).
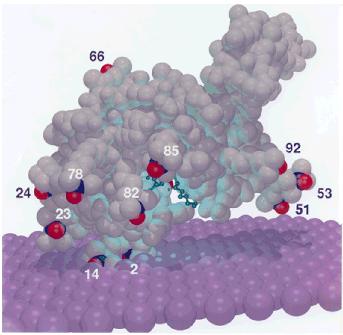
Bee venom phospholipase A2 on a membrane surface providing an experimental basis for the mechanism by which the venom disrupts membranes.
