Chapter 18 Apriori and Post-Hoc Comparisons
This chapter is about how to test hypotheses on data from ANOVA designs that are more specific than the omnibus test which just tests if the means are significantly different from each other. Examples include comparing just two of the means, or comparing one mean (e.g. a control condition) to all of the other means.
The main issue here is familywise error, discussed in the last chapter, which is the fact that the probability of making one or more Type I errors increases with the number of hypothesis tests you make. For example, if you run 10 independent hypothesis tests on your results, each with an alpha value of 0.05, the probability of getting at least one false positive would be:
\[1-(1-0.05)^{10} = 0.401\]
This number, 0.4013, is called the ‘familywise error rate’ or FWER and is clearly unacceptably high. The methods described in this chapter cover the various ways to control, or correct for, familywise error. The more tests you run, the greater the FWER.
Specific hypothesis tests on ANOVA data fall into two categories, ‘A Priori’ and ‘post-hoc’.
A Priori tests are hypothesis tests that you planned on running before you started your experiment. Since there are many possible tests we could make, setting aside a list of just a few specific A Priori tests lets us correct for a much lower familywise error rate.
Post-hoc tests are hypothesis tests that you run after looking at your data. For example, you might want to go back and see if there is a significant difference between the highest and lowest means. Under the null hypothesis, the probability of rejecting a test on the most extreme difference between means will be much greater than \(\alpha\). Or, perhaps we want to go crazy and compare all possible pairs of means. Since there are many tests that you could have run, even if you only pick a few, you need to correct for a larger FWER for post-hoc tests.
18.1 One-Factor ANOVA Example:
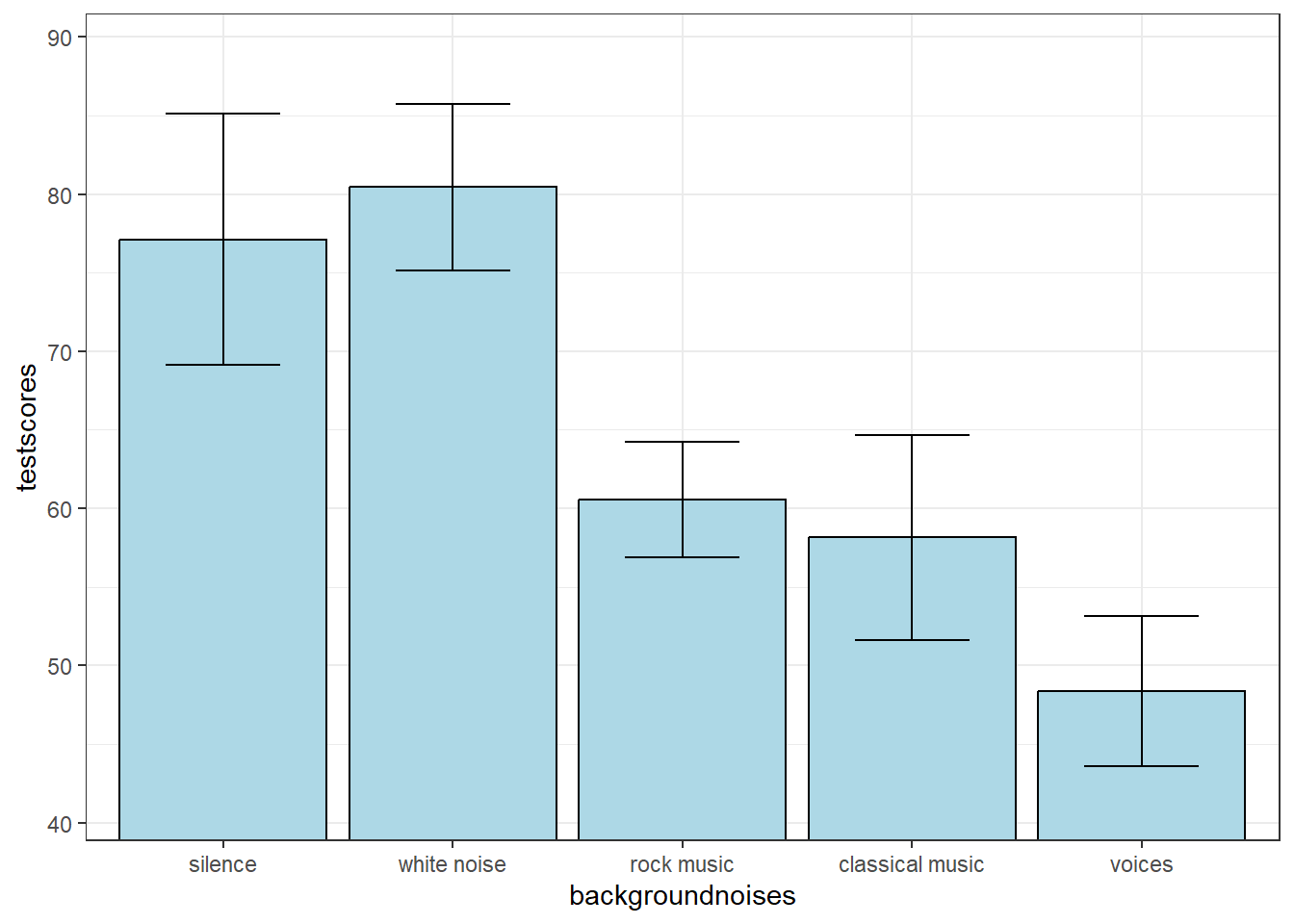
We’ll go through A Priori and post-hoc tests with an example. Suppose you want to study the effect of background noise on test score. You randomly select 10 subjects for each of 5 conditions and have them take a standardized reading comprehension test with the following background noise: silence, white noise, rock music, classical music, and voices.
Throughout this chapter we’ll be referencing this same set of data. You can access it yourself at:
http://courses.washington.edu/psy524a/datasets/AprioriPostHocExample.csv
Your experiment generates the following statistics:
| mean | n | sd | sem | |
|---|---|---|---|---|
| silence | 77.11 | 10 | 25.26436 | 7.989291 |
| white noise | 80.45 | 10 | 16.70617 | 5.282955 |
| rock music | 60.56 | 10 | 11.63856 | 3.680435 |
| classical music | 58.15 | 10 | 20.61112 | 6.517809 |
| voices | 48.37 | 10 | 15.04113 | 4.756424 |
The results of the ANOVA are:
| df | SS | MS | F | p | |
|---|---|---|---|---|---|
| Between | 4 | 7285.217 | 1821.3042 | 5.3445 | p = 0.0013 |
| Within | 45 | 15335.044 | 340.7788 | ||
| Total | 49 | 22620.261 |
Here’s a plot of the means with error bars as the standard error of the mean:
18.2 A Priori Comparisons
An A Priori test is a hypothesis test that you had planned to make before you conducted the experiment. They’re sometimes called planned comparisons.
18.2.1 t-test for two means
The simplest A Priori tests is a comparison between two means. In our example, suppose before we ran the experiment we had the prior hypothesis that there is a difference in mean test score between the silence and the voices conditions. This leads to comparing the means \(\overline{X}_{1} = 77.11\) and \(\overline{X}_{5} = 48.37\)
You’d think that we would simply conduct an independent measures two-tailed t-test using these two group means and variances, while ignoring all of the other conditions (and sometimes this is what people do). But since we have \(MS_{within}\), we should use this value since it’s a better estimate of the population variance than the pooled variance from just two groups (assuming homogeneity of variance).
The old t-statistic was (since we have equal sample size, n):
\[t=\frac{\overline{X}_{1}-\overline{X}_{5}}{\sqrt{\frac{s_{1}^2+s_{5}^2}{n}}} = \frac{77.11 - 48.37}{\sqrt{\frac{25.3^2 + 15^2}{10}}} = 3.091 \]
With \(df = (n-1)+(n-1) = 2n-2 = 18\).
However, we have mean-squared error within (\(MS_w\)) from the ANOVA, which is a better estimate of our population variance than \(s_{1}^2\) and \(s_{5}^2\). So we’ll use it instead of the pooled variance:
\[t= \frac{\overline{X}_{1}-\overline{X}_{5}}{\sqrt{\frac{2MS_{w}}{n}}}\]
The degrees of freedom is now N-k, since this is the df for \(SS_{within}\).
For our example:
\[t= \frac{77.11 - 48.37}{\sqrt{\frac{(2)340.8}{10}}} = 3.4813\]
With df =N-k = 50 - 5 = 45, the p-value of t is 0.0011.
Since we planned on making this comparison ahead of time, and this is our only A Priori comparison, we can use this test to reject \(H_{0}\) and say that there is a difference in the mean test score between the silence and the voices conditions.
As an exercise, make a planned comparison t-test between the rock music and classical music conditions. You should get a t-value of 0.2919 and a p-value of 0.7717.
18.2.2 ‘Contrast’ for two means
Another way of thinking about the comparison we just made between the means from the silence and the voices conditions is to consider the numerator of our t-test as a ‘linear combination’ of means. A linear combination is simply a sum of weighted values. For this comparison, we assign a weight of 1 for the silence condition and a weight of -1 for the voices condition. All other means get zero weights. We use a lower case ‘psi’ (\(\psi\)) to indicate this weighted sum of means, with weights \(a_{i}\) for each mean, \(\overline{X}_{i}\). For this example:
\[\psi = (1)(77.11) + (-1)(48.37) = 28.7400\]
The hypothesis test for contrasts can be done as either a t or an F test since when \(df_{bet}\) =1, F = \(t^{2}\). We’ll use the F test in this chapter. The numerator of the F test is calculated with the following sums of squared error:
\[SS_{contrast} = \frac{{\psi}^2}{\sum{{(a_i^2/n_i)}}}\]
For equal sample sizes, like this example, this simplifies to:
\[SS_{contrast} = \frac{{\psi}^2}{(\sum{{a_i^2})/n}}\]
Which for our example is:
\[SS_{contrast} =\frac{28.74^2}{(1^2+(-1)^2)/10} = 4129.94\]
The mean squared error is always the sum of squared error divided by the degrees of freedom. The df for A Priori contrasts is always 1, so the numerator of the F test will be:
\[MS_{contrast} = \frac{SS_{contrast}}{1} = SS_{contrast}\]
The denominator of the F test for A Priori contrasts is the same denominator as for the omnibus F, or \(MS_{w}\). So our F value is:
\[F(1,df_{within}) = \frac{MS_{contrast}}{MS_{within}}\]
Which for our example is:
\[F(1,45) = \frac{4129.94}{340.78} = 12.12\]
The p-value for this value of F is 0.0011, which is the same p-value as for the t-test above. That’s because our F statistic is equal to \(t^{2}\) (\(12.12 = 3.48^{2}\)).
Note that our contrast weights, 1 and -1, can vary by a scale factor. If we used, for example, \(\frac{1}{2}\) and \(-\frac{1}{2}\) we’d get the same F statistic and p-value. But the contrasts do have to add up to zero in order to test the null hypothesis that the population means are the same.
18.2.3 Contrast for groups of means
Contrasts also allow us to compare groups of means with other groups of means. In our example, suppose we have the prior hypothesis that music in general has a different effect on test scores than white noise. That is, we want to compare the average of the two music conditions (rock and classical) with the white noise condition.
Our weights will be 1 for the white noise condition, and -.5 for the rock and -.5 for the classical conditions, and zero for the remaining conditions. The corresponding linear combination of means is:
\[\psi = (1)(80.45) + (-0.5)(60.56) + (-0.5)(58.15) = 21.0950\]
You should convince yourself that this value, \(\psi\), is the difference between the white noise condition and the average of the two music conditions. It should have an expected value of zero for the null hypothesis because the weights add up to zero.
The mean squared error for this contrast is:
\[SS_{contrast} = \frac{{\psi}^2}{(\sum{{a_i^2})/n}} = \frac{21.095^2}{\frac{(1)^2 +(-0.5)^2+(-0.5)^2}{10}} = 2966.66\]
As always for contrasts, \(df_{contrast} = 1\), so \(MS_{contrast} = \frac{SS_{contrast}}{df_{contrast}} = SS_{contrast}\)
so our F statistic is:
\[F(1,45) = \frac{2966.66}{340.78} = 8.71\]
The p-value for this value of F is 0.005
18.2.3.1 Orthogonal contrasts and independence
We have now made two contrasts. We just compared the effects of white noise to the average of the effects of rock and classical music on test score. Before that we compared the silence and voices conditions. You should appreciate that these two contrasts are independent simply because they don’t share any groups in common.
Contrasts can be independent even if they share groups. Formally, two contrasts are independent if the sum of the products of their weights (the ‘dot product’) add up to zero. When this happens, the two contrasts are called orthogonal. In our example:
c1 = [1, 0, 0, 0, -1]
and our new contrast is
c2 = [0, 1, -1/2, -1/2, 0]
The sum of their products is 0:
(1)(0) + (0)(1) + (0)(-.5) + (0)(-.5) + (-1)(0) = 0
Another contrast that is orthogonal to the second one is:
c3 = [0, 0, 1, -1, 0]
This is because (0)(0) + (1)(0) + (-.5)(1) + (.5)(1) + (0)(0) = 0
What does this contrast test? It compares the test scores for the rock and classical music conditions. Notice that this contrast is also orthogonal to c1, the first contrast.
It turns out that there are exactly as many mutually orthogonal contrasts as there are degrees of freedom for the numerator of the omnibus (k-1). So there should be 4 orthogonal contrasts for our example (though this is not a unique set of 4 orthogonal contrasts). This leaves one more contrast. Can you think of it?
The answer is:
c4 = [1/2 -1/3 -1/3 -1/3 1/2]
Show that c4 is orthogonal to the other three. What is it comparing? It’s the mean of the silence and voices conditions compared to the mean of the other three conditions (white noise, rock and classical). We probably wouldn’t have had an A Priori hypothesis about this particular contrast.
Notice that since each contrast has one degree of freedom, the sum of degrees freedom across all possible contrasts is equal to the degrees of freedom of the omnibus. Likewise, it turns out that the sums of the \(SS_{contrast}\) across all orthogonal contrasts adds up to the \(SS_{bet}\).
If two contrasts are orthogonal, then the two tests are ‘independent’. If two tests are independent, then the probability of rejecting one test does not depend of the probability of rejecting the other. An example of two contrasts that are not independent is comparing silence to white noise for the first contrast, and silence to rock music for the second contrast. You should see that if we happen to sample an extreme mean for the silence, then there will a high probability that both of the contrasts will be statistically significant. Even though both have a Type I error rate of \(\alpha\), there is will be a positive correlation between the probability of rejecting the two tests.
Testing orthogonal contrasts on the same data set is just like running completely separate experiments. Since orthogonal contrasts are independent, we can easily calculate the familywise error rate:
\[FWER = 1-(1-\alpha)^{n}\]
where \(n\) is the number of orthogonal contrasts.
If a set of tests are not independent, the familywise error rate still increases with the number of tests, but in more complicated ways that will be dealt with in the post-hoc comparison section below.
18.3 Contrasts with R
R doesn’t have any libraries to conduct contrasts, but it’s not too difficult to do them ‘by hand’. I’ve supplied some code to do it for you. First, though, lets’ load in the data set that we’ve been working with and compute the ANOVA. Although it’s a bit lengthy, we’ve covered this in the chapter on ANOVA as sums of squares:
# choose your alpha
alpha <- .05
# load in the data
mydata<-read.csv("http://www.courses.washington.edu/psy524a/datasets/AprioriPostHocExample.csv")
# set the background noise levels as a 'factor' in a specific order
mylevels <- unique(mydata$backgroundnoises)
mylevels <- factor(mylevels,levels = mylevels[c(5,3,2,1,4)])
mydata$backgroundnoises <- factor(mydata$backgroundnoises,levels = mylevels)
# run the ANOVA
anova.out <- anova(lm(testscores ~ backgroundnoises,data=mydata))
# pull out values from the anova summary
dfbet <- anova.out$Df[1]
SSbet <- anova.out$`Sum Sq`[1]
MSbet <- anova.out$`Mean Sq`[1]
dfw <- anova.out$Df[2]
SSw <- anova.out$`Sum Sq`[2]
MSw <- anova.out$`Mean Sq`[2]
F.value <- anova.out$`F value`[1]
p.value <- anova.out$`Pr(>F)`[1]
# generate a data frame containing statistics for each level
mydata.summary <- data.frame(
levels = mylevels,
mean = tapply(mydata$testscores,mydata$backgroundnoises,mean),
n = tapply(mydata$testscores,mydata$backgroundnoises,length),
sd = tapply(mydata$testscores,mydata$backgroundnoises,sd))
mydata.summary$sem <- mydata.summary$sd/sqrt(mydata.summary$n)
# re-order the rows
mydata.summary <- mydata.summary[c(5,3,2,1,4),]
mydata.summary## levels mean n sd sem
## silence silence 77.11 10 25.26436 7.989291
## white noise white noise 80.45 10 16.70617 5.282955
## rock music rock music 60.56 10 11.63856 3.680435
## classical music classical music 58.15 10 20.61112 6.517809
## voices voices 48.37 10 15.04113 4.756424Now that we have all of our ANOVA results stored in variables, we’re ready to compute contrasts. The coefficients for the four contrasts will be stored in a 4x5 matrix (4 rows by 5 columns since there are four contrasts and 5 ‘background noise’ levels):
# Contrasts
contrast <- matrix(c(
1, 0, 0, 0, -1,
0, 1, -0.5, -0.5, 0,
0, 0, 1, -1, 0,
0.5, -0.33333, -0.33333, -0.33333, 0.5),nrow = 4, byrow = TRUE)We’re now ready to do the math to compute \(\psi\), \(SS_{contrast}\) and the corresponding F-statistics and p-values. I’m not expecting you to be able to program this yourself - but it’s yours to have and will work on any set of contrasts that you define yourself. If your curious, the %*% means ‘element by element’ multiplication (like ’.*’ in Matlab if that helps) which is used to calculate \(\psi\).
psi <- contrast %*%mydata.summary$mean
SScontrast <- psi^2/colSums(t(contrast^2)/as.vector(mydata.summary$n))
Fcontrast <- SScontrast/MSw
pcontrast <- 1-pf(Fcontrast,1,dfw)
contrast.result = data.frame(contrast,psi,round(SScontrast,4),round(Fcontrast,2),round(pcontrast,4))
colnames(contrast.result) <- c( rownames(mydata.summary),"psi","SS","F","p")
row.names(contrast.result) <- sprintf('c%d',1:nrow(contrast))The resulting table contrast.result looks like this:
| silence | white noise | rock music | classical music | voices | psi | SS | F | p | |
|---|---|---|---|---|---|---|---|---|---|
| c1 | 1.0 | 0.00 | 0.00 | 0.00 | -1.0 | 28.740 | 4129.9380 | 12.12 | 0.0011 |
| c2 | 0.0 | 1.00 | -0.50 | -0.50 | 0.0 | 21.095 | 2966.6602 | 8.71 | 0.0050 |
| c3 | 0.0 | 0.00 | 1.00 | -1.00 | 0.0 | 2.410 | 29.0405 | 0.09 | 0.7717 |
| c4 | 0.5 | -0.33 | -0.33 | -0.33 | 0.5 | -3.646 | 159.5213 | 0.47 | 0.4974 |
18.3.1 Breaking down \(df_{between}\) with \(SS_{contrast}\)
For an ANOVA with \(k\) groups there will be \(k-1\) independent contrasts. These contrasts are not unique - there can be multiple sets of \(k-1\) orthogonal contrasts. But for any set, it turns out that \(SS_{between}\) is the sum the \(k-1\) \(SS_{contrast}\) values:
\[4129.94+2966.66+29.0405+159.578 = 7285.22 = SS_{between}\]
The intuition behind this is that the 4 contrasts are breaking down the total amount of variability between the means (\(SS_{between}\)) into separate independent components, each producing their own hypothesis test.
18.4 Controlling for familywise error rates
The most common way to control for familywise error is to simply lower the alpha value when making multiple independent comparisons. This comes at the expense of lowering the power for each individual test because, remember, decreasing alpha decreases power.
There is a variety and growing number of correction techniques, we’ll cover just a few here.
18.4.1 Bonferroni correction
Bonferroni correction is the easiest, oldest, and most common way to correct for FWER. All you do is reduce alpha by dividing it by the number of comparisons. For example, if you want to make 4 comparisons and want the FWER to be below 0.05, you simply test each comparison with an alpha value of 0.05/4 = 0.0125.
Our contrasts produced p-values of 0.0011, 0.005, 0.7717, and 0.4973. If we were to make all four A Priori comparisons, we’d need to adjust alpha to be 0.05/4 = 0.0125.
We’d therefore reject the null hypothesis for contrasts 1 and 2 but not for contrasts 3 and 4.
18.4.2 Šidák correction
Some software packages correct for familywise error using something called the Šidák correction. The result is almost exactly the same as the Bonferroni correction, but it’s worth mentioning here so you know what it means when you see a button for it in software packages like SPSS.
Remember, the familywise error rate is the probability of making one or more false positives. The Bonferroni correction is essentially assuming that the familywise error rate grows in proportion to the number of comparisons so we scale alpha down accordingly. But we know that the family wise error rate is \(1-(1-\alpha)^m\), where m is the number of comparisons.
For example, for a Bonferroni correction with \(\alpha\) = .05 and 4 comparisons, we need to reduce the alpha value for each comparison to 0.05/4 = 0.0125. The familywise error rate is now
\[1-(1-0.0125)^4 = 0.049\]
It’s close to 0.05 but it’s just a little lower. To bring the FWER up to exactly 0.05 we need to use:
\[\alpha' = 1-(1-\alpha)^{1/m}\]
With 4 comparisons and \(\alpha\) = 0.05, the adj alpha is:
\[\alpha' = 1-(1-0.05)^{1/4} = 0.01274\]
So instead of using an alpha of 0.0125 you’d use 0.01274. The difference between the Šidák correction and the Bonferroni correction is minimal but technically the Šidák correction sets the probability of getting one or more one false alarms to exactly alpha for independent tests.
18.4.3 Holm-Bonferroni Multistage Procedure
The Holm-Bonferroni procedure is a more forgiving way to correct for familywise error, and has been used more recently, especially for large number of comparisons. The procedure is best described by example. First, we rank-order our p-values from our multiple comparisons in from lowest to highest. From our four contrasts: 0.0011, 0.005, 0.4973, and 0.7717 for contrast numbers 1, 2, 4, and 3 respectively.
We start with the lowest p-value and compare it to the alpha that we’d use for the Bonferroni correction (\(\frac{0.05}{4} = 0.0125\)).
If our lowest p-value is less than this adjusted alpha, then we reject the hypothesis for this contrast. If we fail to reject then we stop. For our example, p = 0.0011 is less than 0.0125, so we reject the corresponding contrast (number 1) and move on.
We then compare or next lowest p-value to a new adjusted alpha. This time we divide alpha by 4-1=3 to get \(\frac{0.05}{3} = 0.0167\), a less conservative value. If we reject this contrast, the we move on to the next p-value and the next adjusted alpha \(\frac{0.05}{2}=0.025\)).
This continues until we fail to reject a comparison, and then we stop.
The idea is that if you manage to reject with the lowest p-value using the full Bonferroni correction for m tests (\(\frac{\alpha}{m}\)), then you can move on to the next p-value and correct only for the remaining m-1 tests (\(\frac{\alpha}{m-1}\)), and so on.
There’s not a clean package to do this in R, but here’s how you can do it on your own:
# rank order the p-values from lowest to highest
pvalue.order <- order(contrast.result$p)
# set up the conditions for terminating the 'while' loop
failed.yet <- FALSE
i <- 1
while (i<=m && !failed.yet){
current.alpha = alpha/(m-i+1)
if (contrast.result$p[pvalue.order[i]]<current.alpha) {
cat(sprintf("contrast %d: Reject H0, F(1,%d) = %5.4f, p = %5.4f < %5.4f\n",
pvalue.order[i],dfw,contrast.result$F[pvalue.order[i]],
contrast.result$p[pvalue.order[i]],current.alpha))
} else {
cat(sprintf("contrast %d: Fail to reject H0, F(1,%d) = %5.4f, p = %5.4f > %5.4f\n",
pvalue.order[i],dfw,contrast.result$F[pvalue.order[i]],
contrast.result$p[pvalue.order[i]],current.alpha))
failed.yet <- TRUE
cat("done, fail to reject the rest.\n")
}
i <- i+1
}## contrast 1: Reject H0, F(1,45) = 12.1200, p = 0.0011 < 0.0125
## contrast 2: Reject H0, F(1,45) = 8.7100, p = 0.0050 < 0.0167
## contrast 4: Fail to reject H0, F(1,45) = 0.4700, p = 0.4974 > 0.0250
## done, fail to reject the rest.This isn’t a programming class, so I don’t expect you to fully understand the code. But it will work for your own set of contrasts. If you’re interested, however, see if you can follow how it works. It uses a while loop, which continues while the condition i<=m && !failed.yet is true. m is the number of contrasts, and i is an index that starts at 1 and increments after each rejected test. Each time through,the contrasts, ranked by their p-values, are compared to alpha/(m-i+1) which starts out at alpha/m for the first contrast, alpha/(m-1) for the second, etc.
The variable failed.yet is a logical (TRUE/FALSE) that starts out as FALSE and turns to true after the first contrast fails to reject. So !failed.yet starts out as true, allowing the while loop to continue until the first fail to reject, or until we run out of contrasts.
Multistage procedures like the Holm-Bonferroni are less conservative and therefore more powerful than the standard Bonferroni correction. They are less widely used probably because they’re more complicated. But as you’ve seen, computers can easily do these things for us.
There are other variants of this sort of multistage procedure, including sorting from highest to lowest p-values, and using a Sidac correction for each test instead of a Bonferroni correction. They all produce similar results and the field has not settled on one procedure in particular.
18.5 Post Hoc Comparisons
Now let’s get more exploratory and make some comparisons that we didn’t plan on making in the first place. These are called post hoc comparisons. For A Priori comparisons, we only needed to adjust for the FWER associated with the number of planned comparisons. For post hoc comparisons, we need to adjust to not just the comparisons we feel like making, but for all possible comparisons of that type that we could have made (e.g all possible pairwise comparisons or all possible contrasts). As you might expect, the more possible tests that you correct for, the more difficult it is to reject, and therefore the lower the power. The following post hoc tests test for increasing numbers of possible tests, and therefore decreasing levels of power.
18.6 Dunnett’s Test for comparing one mean to all others
This is a post-hoc test designed specifically for comparing all means to a single control condition. For our example, it makes sense to compare all of our conditions to the silence condition, giving us 4 tests. Consider the set of contrasts needed for these tests - they are not independent. So we need to do something more fancy than a simple Bonferroni correction.
Dunnett’s test relies on finding critical values from a distribution that is related to the t-distribution called the Dunnett’s t Statistic. It’s easy to run Dunnett’s test in R with the function DunnetTest which requires the ‘DescTools’ library.
Let’s jump straight to the test, since it’s not likely that you’ll ever need to calculate p-values by hand.
With the silence condition as the control condition, we’ll compare the other four means to this one mean (\(\overline{X}_1 = 77.11)\)
Here’s how it works:
dunnett.out <- DunnettTest(testscores ~ backgroundnoises,
data = mydata,control = "silence")
dunnett.out##
## Dunnett's test for comparing several treatments with a control :
## 95% family-wise confidence level
##
## $silence
## diff lwr.ci upr.ci pval
## classical music-silence -18.96 -39.86108 1.941085 0.0852 .
## rock music-silence -16.55 -37.45108 4.351085 0.1563
## white noise-silence 3.34 -17.56108 24.241085 0.9831
## voices-silence -28.74 -49.64108 -7.838915 0.0040 **
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The table itself sits in the output under the field having the name of the control condition as a ‘matrix’. It’s useful to turn it into a data frame. For our example, use this:
Here’s the table formatted so that the significant values are in red:
| diff | lwr | upr | p adj | |
|---|---|---|---|---|
| classical music-silence | -18.96 | -39.8611 | 1.9411 | 0.0852 |
| rock music-silence | -16.55 | -37.4511 | 4.3511 | 0.1563 |
| white noise-silence | 3.34 | -17.5611 | 24.2411 | 0.9831 |
| voices-silence | -28.74 | -49.6411 | -7.8389 | 0.004 |
These p-values are all adjusted for familywise error, so no further correction is needed.
18.7 The Tukey Test
The Tukey Test is a way of correcting for FWER when testing all possible pairs of means. For our example there are 10 pairs of means to test. But like for Dunnett’s test, you can’t use a Bonferroni correction and divide alpha by 10 because not all comparisons are independent. Note that the 4 tests that we ran with Dunnet’s test are a subset of the 10 tests that we’ll be running with the Tukey Test.
The test is based on the distribution that is expected when you specifically compare the largest and smallest mean. If the null hypothesis is true, the probability of a significant t-test for these most extreme means will be quite a bit greater than \(\alpha\), which is the probability of rejecting any random pair of means.
The way for correcting for this inflated false positive rate when comparing the most extreme means is to use a statistic called the Studentized Range Statistic and goes by the letter q.
The statistic q is calculated as follows:
\[q = \frac{\overline{X}_l-\overline{X}_s}{\sqrt{\frac{MS_{within}}{n}}}\]
Where \(\overline{X}_{l}\) is the largest mean, \(\overline{X}_{s}\) is the smallest mean, and n is the sample size of each group (assuming that all sample sizes are equal).
The q-statistic for our most extreme pairs of means is:
\[q = \frac{80.45 - 48.37} {\sqrt{\frac{340.7788}{10}}} = 5.4954\]
The q statistic has its own distribution. Like the t-statistic, it is broader than the normal distribution for small degrees of freedom. Also, q increases in width with increasing number of groups. That’s because as the number of possible comparisons increases, the difference between the extreme means increases.
R has the functions ptukey and qtukey to test the statistical significance of this difference of extreme means. ptukey needs our value of q, the number of groups, and \(df_{within}\). For our pair of extreme means:
## [1] 0.002925262Importantly, even though we selected these two means after running the experiment, this p-value accounts for this bias.
In fact, almost magically, we can use this procedure to compare any or all pairs of means, and we won’t have to do any further correction for multiple comparisons.
18.7.1 Tukey’s ‘HSD’
Back in the chapter on effect sizes and confidence intervals we discussed how an alternative way to make a decision about \(H_{0}\) is to see if the null hypothesis is covered by the confidence interval. The same thing is often done with the Tukey Test.
Instead of finding a p-value from our q-statistic with ptukey, we’ll go the other way and find the value \(q_{crit}\) that covers the middle 95of the q distribution using qtukey. Since it’s a two-tailed test, we need to find q for the top 97.5%
## [1] 4.018417We can go through the same logic that we did when we derived the equation for the confidence interval. If we assume that the observed difference between a pair of means, \(\overline{X}_i-\overline{X}_j\) is the true difference between the means, then we expect that in future experiments, we can expect to find a difference 95% of the time within:
\[\overline{X}_i-\overline{X}_j \pm q_{crit}\sqrt{\frac{MS_{w}}{n}}\] For our example:
\[ q_{crit}\sqrt{\frac{MS_{w}}{n}} = 4.0184\sqrt{\frac{340.7788}{10}} = 23.4579\]
This value, 23.4579 is called Tukeys’ honestly significant difference, or HSD. Any pair of means that differs by more than this amount can be considered statistically significant at the level of \(\alpha\).
The ‘H’ in Tukey’s HSD is for ‘honestly’ presumably because it takes into account all possible pairwise comparisons of means, so we’re being honest and accounting for familywise error.
We can use Tukey’s HSD to calculate a confidence interval by adding and subtracting it from the observed difference between means. Our most extreme means had a difference of
\[80.45 - 48.37 = 32.08\]
The 95% confidence interval for the difference between these means is:
\[(32.08 - 23.4579, 32.08 + 23.4579)\]
which is
\[( 8.6221, 55.5379)\]
The lowest end of this interval is greater than zero, which is consistent with the fact that the p-value for the difference between means (0.0029) is less than \(\alpha\) = 0.05. If the difference between the means is significantly significant (with \(\alpha\) = 0.05), then the 95% confidence interval will never contain zero.
All this is easy to do in R using the tukeyHSD function. There’s just a little hack. It’s an old function which requires the output of an outdated ANOVA function called aov. But you can, instead, send in to tukeyHSD the output of lm after changing it’s class to aov. Here’s how you run the Tukey test on our example. We’ll run the lm function on our data, but instead of passing it into anova like we did before, we’ll change it’s class to ‘aov’ and pass it into TukeyHSD:
lm.out <- lm(testscores ~ backgroundnoises,data =mydata)
class(lm.out) <- c("aov","lm") # total hack
TukeyHSD(lm.out)## Tukey multiple comparisons of means
## 95% family-wise confidence level
##
## Fit: lm(formula = testscores ~ backgroundnoises, data = mydata)
##
## $backgroundnoises
## diff lwr upr p adj
## rock music-classical music 2.41 -21.048015 25.868015 0.9983541
## white noise-classical music 22.30 -1.158015 45.758015 0.0695439
## voices-classical music -9.78 -33.238015 13.678015 0.7599945
## silence-classical music 18.96 -4.498015 42.418015 0.1649044
## white noise-rock music 19.89 -3.568015 43.348015 0.1315002
## voices-rock music -12.19 -35.648015 11.268015 0.5826433
## silence-rock music 16.55 -6.908015 40.008015 0.2803943
## voices-white noise -32.08 -55.538015 -8.621985 0.0029254
## silence-white noise -3.34 -26.798015 20.118015 0.9941624
## silence-voices 28.74 5.281985 52.198015 0.0094156The p-values in the column ‘p adj’ reflect the correction for FWER, so they don’t need to be adjusted to compare to \(\alpha\). Any test for a pair of means with an adjusted p-value less than \(\alpha\) can be rejected.
Here you’ll see adjusted p-values for all possible \(\frac{5 \times 4}{2} = 10\) comparisons. Can you find the row for the biggest difference between means?
You’ll also see the columns ‘lwr’ and ‘upr’. These are the ranges for the 95% confidence interval on the differences between the means. For all cases, if the p-value ‘p adj’ is less than .05, then the 95% confidence interval will not include zero. Is the confidence interval the same as our calculation? It’s off by a \(\pm\) sign flip because tukeyHSD decided to run the test with the opposite order of means (smallest - largest). Otherwise it’s the same, and it doesn’t matter anyway because it’s a two-tailed test. You should notice that the range between ‘lwr’ and ‘upr’ is always the same value of Tukey’s HSD = 23.4579.
18.7.2 Tukey-Kramer Test - for unequal sample sizes
The sample sizes must be equal when using the Tukey test. If the sample sizes varies across groups (called an ‘unbalanced design’) there is a modification of the Tukey test called the ‘Tukey-Kramer method’ which uses a different HSD depending upon which means are being compared.
Specifically, to compare means from group i to group j, with sample sizes \(n_{i}\) and \(n_{j}\) and variances \(s_{i}^{2}\) and \(s_{j}^{2}\), the q-statistic is:
\[ q = \frac{\overline{X}_i-\overline{X}_j}{\sqrt{\frac{ \frac{s_{i}^{2}}{n_{i}} + \frac{s_{j}^{2}}{n_{j}}}{2}}}\] And replace \(df_{within}\) with:
\[df_{ij}= \frac{ \left(\frac{s_{i}^{2}}{n_{i}} + \frac{s_{j}^{2}}{n_{j}}\right)^{2}} { \frac{\left(\frac{s_{i}^{2}}{n_{i}}\right)^{2}}{n_{i}-1} + \frac{\left(\frac{s_{j}^{2}}{n_{j}}\right)^{2}}{n_{j}-1} }\]
It’s kind of ugly, but here’s some R code that makes all possible paired comparisons based on our summary table mydata.summary that we computed above:
k <- nrow(mydata.summary)
# create a matrix of NA's to hold our p-values
pTable <- data.frame(matrix(nrow = k-1,ncol = k-1))
row.names(pTable) <- row.names(mydata.summary)[1:k-1]
colnames(pTable) <- row.names(mydata.summary)[2:k]
for (i in 1:(k-1)) {
for (j in (i+1):k) {
# compare means from group i to group j
ni <- mydata.summary$n[i]
nj <- mydata.summary$n[j]
sni <- mydata.summary$sd[i]^2/ni
snj <- mydata.summary$sd[j]^2/nj
q <- (abs(mydata.summary$mean[i]-mydata.summary$mean[j]))/sqrt((sni+snj)/2)
df <- (sni+snj)^2/(sni^2/(ni-1)+snj^2/(nj-1))
pTable[i,j-1] <- round(1-ptukey(q,k,df),4)
}
}
pTable[is.na(pTable)] <- ""Which produces a nice table. I’ve rendered the table using the kable package, and colored the significant comparisons red.
| white noise | rock music | classical music | voices | |
|---|---|---|---|---|
| silence | 0.9965 | 0.3741 | 0.3844 | 0.0505 |
| white noise | 0.0475 | 0.1027 | 0.0022 | |
| rock music | 0.9974 | 0.2955 | ||
| classical music | 0.7447 |
Even though we ran the Tukey-Kramer test on the same data set, which has equal sample sizes, the p-values aren’t the same as for the regular Tukey Test. Usually, but not always, the Tukey-Kramer test will be less powerful (have larger p-values) than the Tukey Test because it is only using the variance for two means at a time which has a lower \(df\) than for the regular Tukey Test.
18.8 The Sheffe’ Test: correcting for all possible contrasts
The Sheffe’ test allows for post-hoc comparisons across all possible contrasts (including non-orthogonal contrasts). Since there are many more possible contrasts than pairs of means, the Sheffe’ test has to control for more possible comparisons than either Dunnet’s test or the Tukey Test, and is therefore the most conservative and least powerful.
Remember, the F-test for a contrast is conducted by first computing the weighted sum of means:
\[\psi = \sum{a_i\overline{X}_i}\]
The \(SS_{contrast}\) (and \(MS_{contrast}\)):
\[SS_{contrast} = MS_{contrast} =\frac{{\psi}^2}{\sum{{a_i^2/n_i}}}\]
and then the F statistic:
\[F(1,df_{w}) = \frac{MS_{contrast}}{MS_{w}}\]
The Sheffe’ test adjusts for multiple comparisons by multiplying the critical value of F for the original ‘omnibus’ test by k-1 (the number of groups minus one). In our example, the critical value of F for the omnibus test can be calculated using R’s qf function:
## [1] "F_crit <- qf(1-0.05,4,45)"## [1] 2.578739Our adjusted critical value for F is (4)(2.5787) = 10.315. Any contrast with an F-statistic greater than this can be considered statistically significant.
Instead of a critical value of F, we can convert F = 10.315 into a modified value of \(\alpha\). Using R’s pf function we get:
## [1] "alpha_Sheffe <-1-pf(F_crit_Sheffe,1,45)"## [1] 0.002438332Any contrast with a p-value less than 0.0024 is considered statistically significant. This is about 1 in 400 contrasts.
Remember, our four contrasts had weights:
c1 = [1 0 0 0 -1]
c2 = [0 1 -1/2 -1/2 0]
c3 = [0 0 1 -1 0]
c4 = [1/2 -1/3 -1/3 -1/3 1/2]
The F values for these four contrasts are 12.12, 8.71, 0.09, and 0.47, and the four corresponding p-values are 0.0011, 0.005, 0.7717, and 0.4973.
Comparing these F values to our adjusted critical value of F, or comparing the p-values to 0.0024, lets us reject the null hypothesis for contrasts 1 but not for contrasts 2, 3 and 4. How does this compare to the A Priori Bonferroni test we used to compare these contrasts?
With the Sheffe’ test, the door is wide open to test any post-hoc comparison we want. Looking at the bar graph, it looks like there is a difference between the average of the silence, and white noise conditions, and the average of the rock music, classical music, and voices conditions. This would be a contrast with weights:
[1/2, 1/2, -1/3, -1/3, -1/3]
If you work this out you get an F value of 18.7686. This exceeds the Sheffe’-adjusted F-value of 10.315 so we can say that there is a significant difference between these groups of means.
You can also convert the F-statistics from any contrasts to ‘adjusted’ p-values which be compared to \(\alpha = .05\). This is done by simply dividing each contrast’s F-value by (k-1) and passing it through R’s pf function with degrees of freedom from the original ANOVA.
Here’s a modified version of the code above that replaces the regular p-value with Sheffe’s ‘adjusted’ p-value for each contrast. These can be compared directly to your value of \(\alpha\) to determine statistical significance:
contrast = t(data.frame(c( 1, 0, 0, 0, -1),
c( 0, 1,-.5,-.5, 0),
c( 0, 0, 1, -1, 0),
c(3,-2,-2,-2,3)/6,
c(3,3,-2,-2,-2)/6))
psi <- contrast %*%mydata.summary$mean
SScontrast <- psi^2/colSums(t(contrast^2)/as.vector(mydata.summary$n))
Fcontrast <- SScontrast/MSw
pcontrast <- 1-pf(Fcontrast/dfbet,dfbet,dfw) # calculate the 'adjusted' p-value
contrast.result = data.frame(contrast,psi,round(SScontrast,4),round(Fcontrast,2),round(pcontrast,4))
colnames(contrast.result) <- c( rownames(mydata.summary),"psi","SS","F","adj p")
row.names(contrast.result) <- sprintf('c%d',1:nrow(contrast))The resulting table contrast.result (including our 5th made-up contrast) looks like this, with the statistically significant p-values in red:
| silence | white noise | rock music | classical music | voices | psi | SS | F | adj p | |
|---|---|---|---|---|---|---|---|---|---|
| c1 | 1.0 | 0.00 | 0.00 | 0.00 | -1.00 | 28.7400 | 4129.9380 | 12.12 | 0.027 |
| c2 | 0.0 | 1.00 | -0.50 | -0.50 | 0.00 | 21.0950 | 2966.6602 | 8.71 | 0.0869 |
| c3 | 0.0 | 0.00 | 1.00 | -1.00 | 0.00 | 2.4100 | 29.0405 | 0.09 | 0.9991 |
| c4 | 0.5 | -0.33 | -0.33 | -0.33 | 0.50 | -3.6467 | 159.5781 | 0.47 | 0.9758 |
| c5 | 0.5 | 0.50 | -0.33 | -0.33 | -0.33 | 23.0867 | 6395.9301 | 18.77 | 0.003 |
It feels like cheating - that we’re making stuff up. And yes, post-hoc tests are things you made up after you look at the data. I think if feels wrong because we’re so concerned about replicability and preregistration. But with the proper correction for multiple comparisons, this is totally fine. In fact, I’ll bet that most of the major discoveries on science have been made after noticing something interesting in the data that wasn’t anticipated. I would even argue that sticking strictly to your preregistered hypotheses could slow down the progress of science.
18.9 Summary
These notes cover just a few of the many A Priori and post hoc methods for controlling for multiple comparisons. Statistics is a relatively new and developing field of mathematics, so the standards for these methods are in flux. Indeed, SPSS alone provides a confusing array of post-hoc methods and allows you to run many or all of them at once. It is clearly wrong to run a bunch of post-hoc comparisons and then pick the comparison that suits you. This sort of post hoc post hoc analysis can lead to a sort of meta-familywise error rate.
It’s also not OK to treat a post hoc comparison with an A Priori method. You can’t go back and say “Oh yeah, I meant to make that comparison” if it hadn’t crossed your mind.
In the end, all of these methods differ by relatively small amounts. If the significance of your comparisons depend on which specific A Priori or post hoc test you choose, then you’re probably taking the .05 or .01 criterion too seriously anyway.